Data Driven Drug Discovery
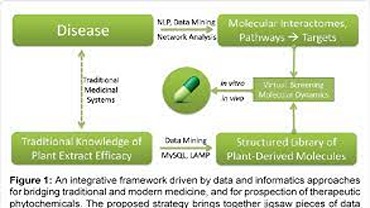
Methodological improvements are imperative to keep up with the need for novel drug molecules that exhibit required properties. For instance, the average time for a drug to reach the market is about 13 years. Thankfully, the improvement in computational power in tandem with advancements in machine learning algorithms has enabled chemists to look at this problem from new paradigms. This area of research identifies various sub parts in the drug discovery pipeline (ex: target identification, lead optimization) where modern frameworks can be used in effective way. These modern methods involve the use of various generative deep learning frameworks, reinforcement learning, transformers et cetera.
Keep Reading...Datasets
Methodological improvements are imperative to keep up with the need for novel drug molecules that exhibit required properties. For instance, the average time for a drug to reach the market is about 13 years. Thankfully, the improvement in computational power in tandem with advancements in machine learning algorithms has enabled chemists to look at this problem from new paradigms. This area of research identifies various sub parts in the drug discovery pipeline (ex: target identification, lead optimization) where modern frameworks can be used in effective way. These modern methods involve the use of various generative deep learning frameworks, reinforcement learning, transformers et cetera.
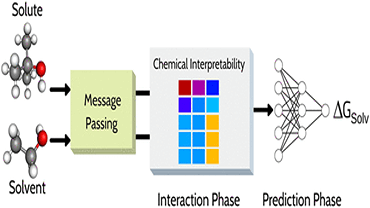
Machine Learning for Chemistry
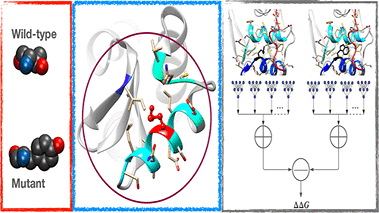
Engineering proteins to have desired properties by mutating amino acids at specific sites is commonplace. Such engineered proteins must be stable to function. Experimental methods used to determine stability at throughputs required to scan the protein sequence space thoroughly are laborious. To this end, many machine learning based methods have been developed to predict thermodynamic stability changes upon mutation. These methods have been evaluated for symmetric consistency by testing with hypothetical reverse mutations
Keep Reading...Molecular Simulations
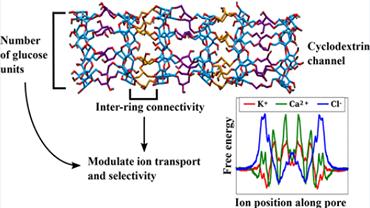
Our group focuses on the protein unfolding dynamics and drug binding to nucleic acid structure by the use of classical molecular dynamics (MD) simulation techniques. This group has worked on understanding the atomistic level details of highly favorable aromatic urea interactions in protein unfolding dynamics, role of such interactions in drug design where urea-moeity based scaffolds can oprove to be viable strong binders. Synthetic ion channels, namely Cyclodextrins have been studied using MD-based free energy calculations. Role of the transmembrane domain topology on the activity of the HIV-1 Vpu virus and ability of antimicrobial agent like cholic acid (CA) derived amphiphiles to combat Gram positive bacterial infections were studied using MD tools.
Keep Reading...Electronic Structure Theory
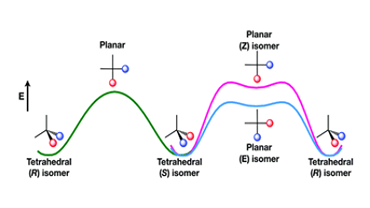
Computational studies of the mechanics and dynamics of small molecule can unravel the mechanism of complex chemical reactions. Our group has studied oxidation of greenhouse gas CO on metal cluster, benchmarked various quantum mechanical methods to model small chemical reactions and has studied the role of basis set to capture molecular properties in cyclic systems. Electronic structure analysis aid in understanding experimental results, and our group has studied photocatalytic reactions, functionalization of cyclic systems, in collaboration with experimental groups. Our group also focuses on the dynamics of tetrahedral centres to mark the first study of stereochemical inversion without bond breaking.
Keep Reading...AI for Healthcare
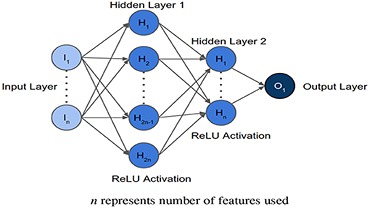
Healthcare institutions have access to a large repository of medical records, which can reduce the time taken for an initial diagnosis/prognosis if leveraged efficiently and ethically. We use and build upon diverse Deep Learning models to improve current pathology identification/localization approaches across different modalities like CTs, X-rays, MRIs, ECGs, etc. Given the variability of healthcare data, class imbalances, dataset shift, etc., we also focus on self-supervised learning approaches and various explainability methods.
Keep Reading...