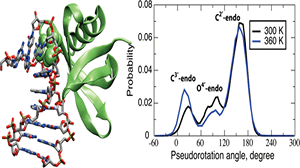
Sac7d belongs to a family of chromosomal proteins, which are crucial for thermal stabilization of DNA at higher growth temperatures. It is capable of binding DNA nonspecifically, and is responsible for the increase in the melting temperature of DNA in the bound form up to 85 °C. Molecular dynamics (MD) simulations were performed at different temperatures on two protein−DNA complexes of Sac7d. Various structural and energetic parameters were calculated to examine the DNA stability and to investigate the conformational changes in DNA and the protein−DNA interactions. Room temperature simulations indicated very good agreement with the experimental structures. The protein structure is nearly unchanged at both 300 and 360 K, and only up to five base pairs of the DNA are stabilized by Sac7d at 360 K. However, the MD simulations on DNA alone systems show that they lose their helical structures at 360 K further supporting the role of Sac7d in stabilizing the oligomers. At higher temperatures (420 and 480 K), DNA undergoes denaturation in the presence and the absence of the protein. The DNA molecules were found to undergo B- to A-form transitions consistent with experimental studies, and the extent of these transitions are examined in detail. The extent of sampling B- and A-form regions was found to show temperature and sequence dependence. Multiple MD simulations yielded similar results validating the proposed model. Interaction energy calculations corresponding to protein−DNA binding indicates major contribution due to DNA backbone, explaining the nonspecific interactions of Sac7d.

UD Priyakumar, G Harika, G Suresh