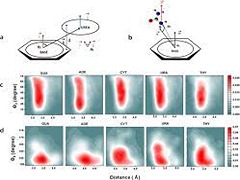
Understanding the structure-function relationships of RNA has become increasingly important given the realization of its functional role in various cellular processes. Chemical denaturation of RNA by urea has been shown to be beneficial in investigating RNA stability and folding. Elucidation of the mechanism of unfolding of RNA by urea is important for understanding the folding pathways. In addition to studying denaturation of RNA in aqueous urea, it is important to understand the nature and strength of interactions of the building blocks of RNA. In this study, a systematic examination of the structural features and energetic factors involving interactions between nucleobases and urea is presented.

T Jaganade, A Chattopadhyay, NM Pazhayam, UD Priyakumar.