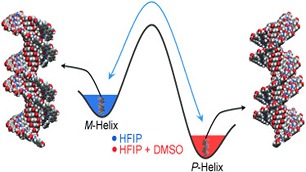
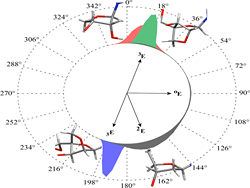
Mastering the handedness: Understanding the roles of various parameters in orchestrating the preferential chiral molecular organization in supramolecular self-assembly processes is of great significance in designing novel molecular functional systems. This study emphasizes the role of cyclic dipeptide chiral auxiliaries on the solvent-induced helical assembly and reversible chiroptical switching of naphthalenediimides (see figure: HFIP=1,1,1,3,3,3-hexafluoroisopropanol).

S Manchineella, V Prathyusha, UD Priyakumar, T Govindaraju,