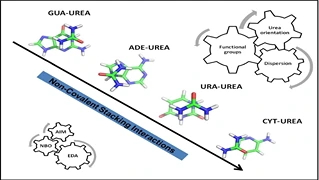
Urea-assisted denaturation of protein and RNA has been shown to be a valuable tool to study their stabilities and folding phenomena. It has been shown that stacking interactions between nucleobases and urea are one of the driving forces of denaturation. In this study, the ability of urea to form unconventional stacking interactions with RNA bases is investigated by performing high-level quantum calculations (RI-MP2/aug-cc-pVDZ level) on a few thousands of model systems. Four systems were considered based on the RNA nucleobases (GUA, ADE, CYT, and URA) for the investigation.

N Alodia, T Jaganade, UD Priyakumar,